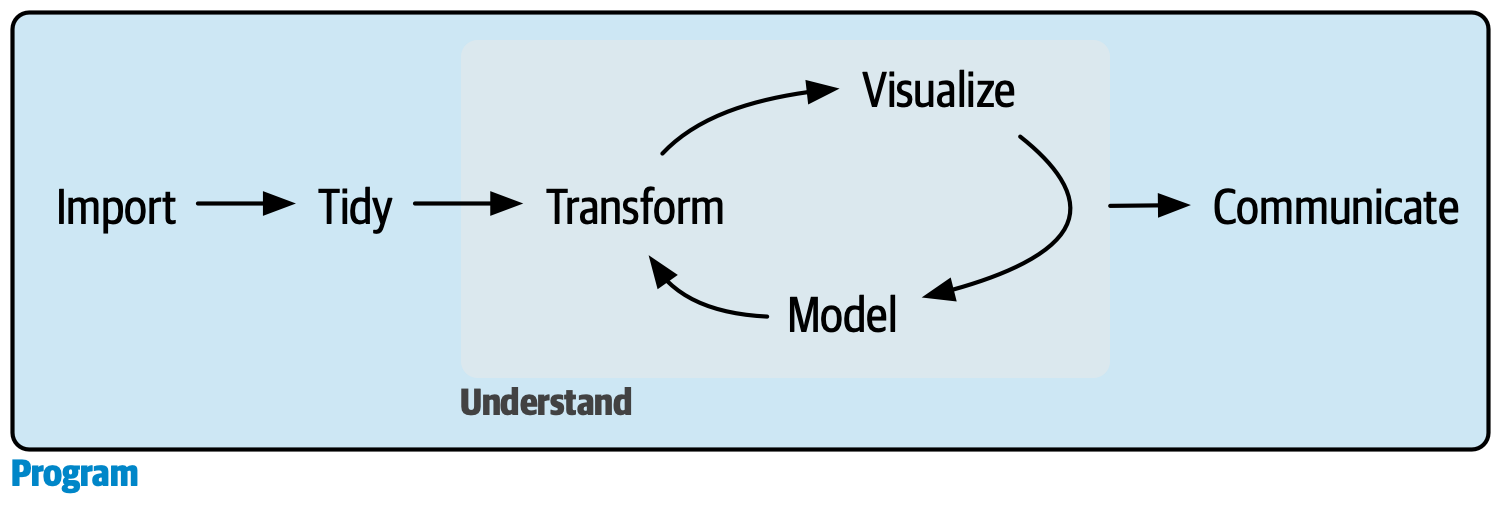
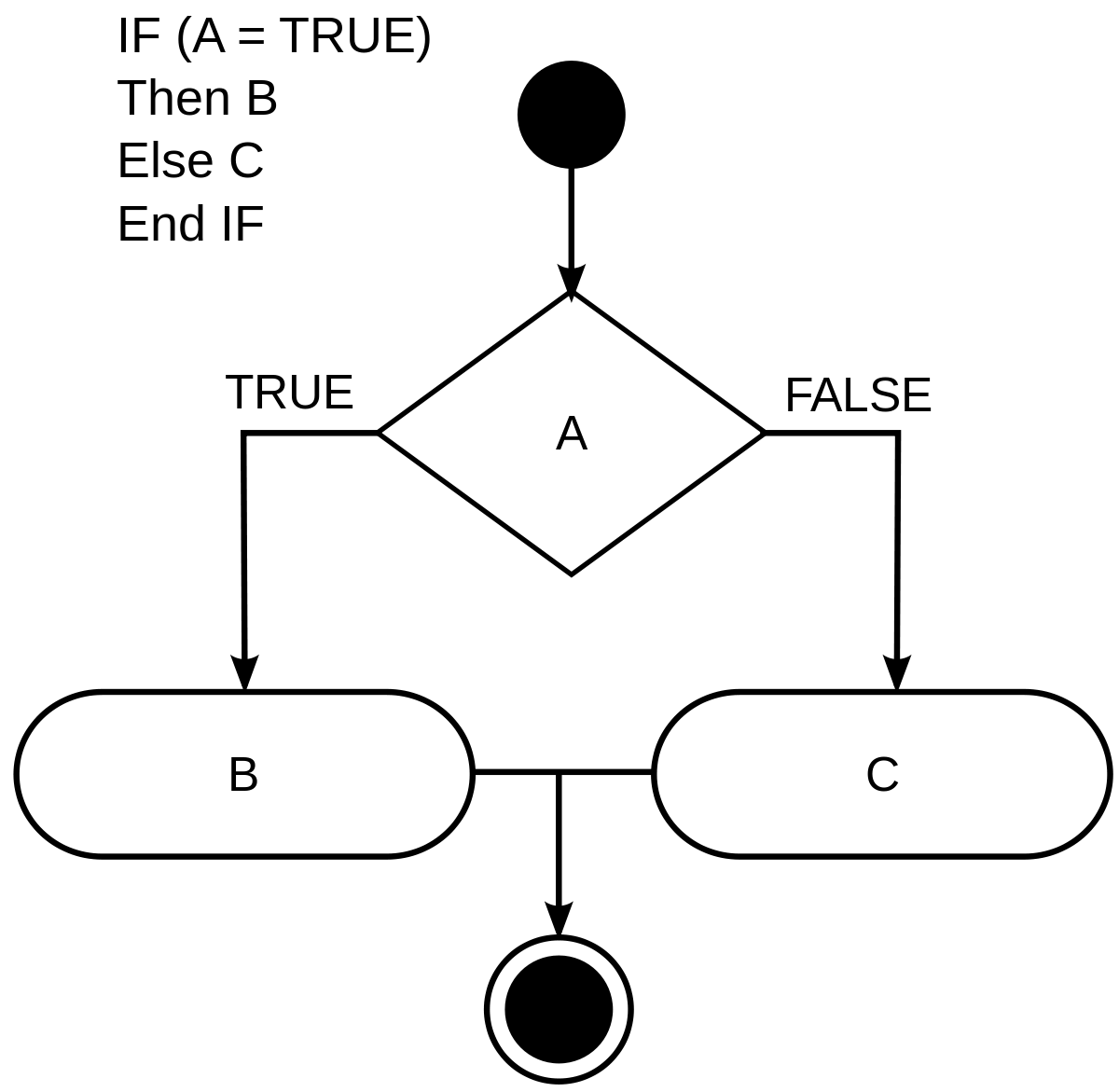
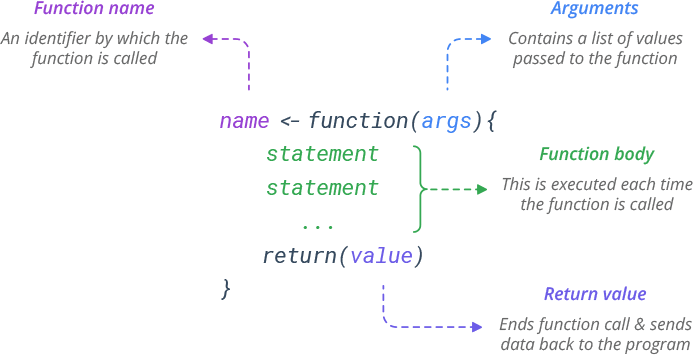
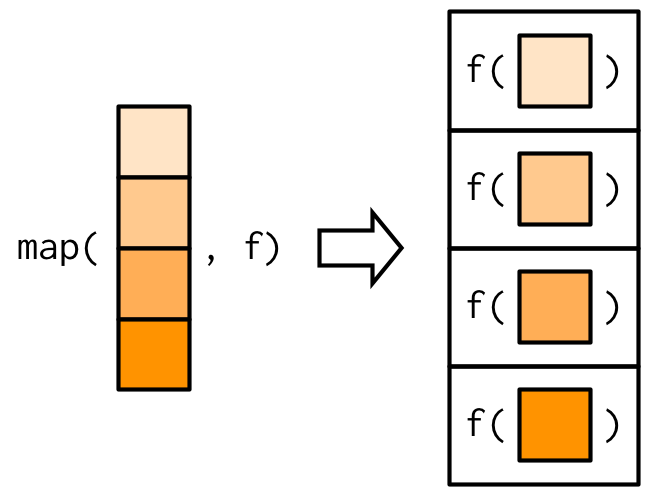
Main takeaways
map()is more readable, faster, and easily extendable with other data science tasks (e.g. wrangling and visualisation) using%>%purrr::map()is simpler to writeThere is one function for each type of output:
map()makes a listmap_lgl()makes a logical vectormap_int()makes an integer vectormap_dbl()makes a double vectormap_chr()makes a character vector